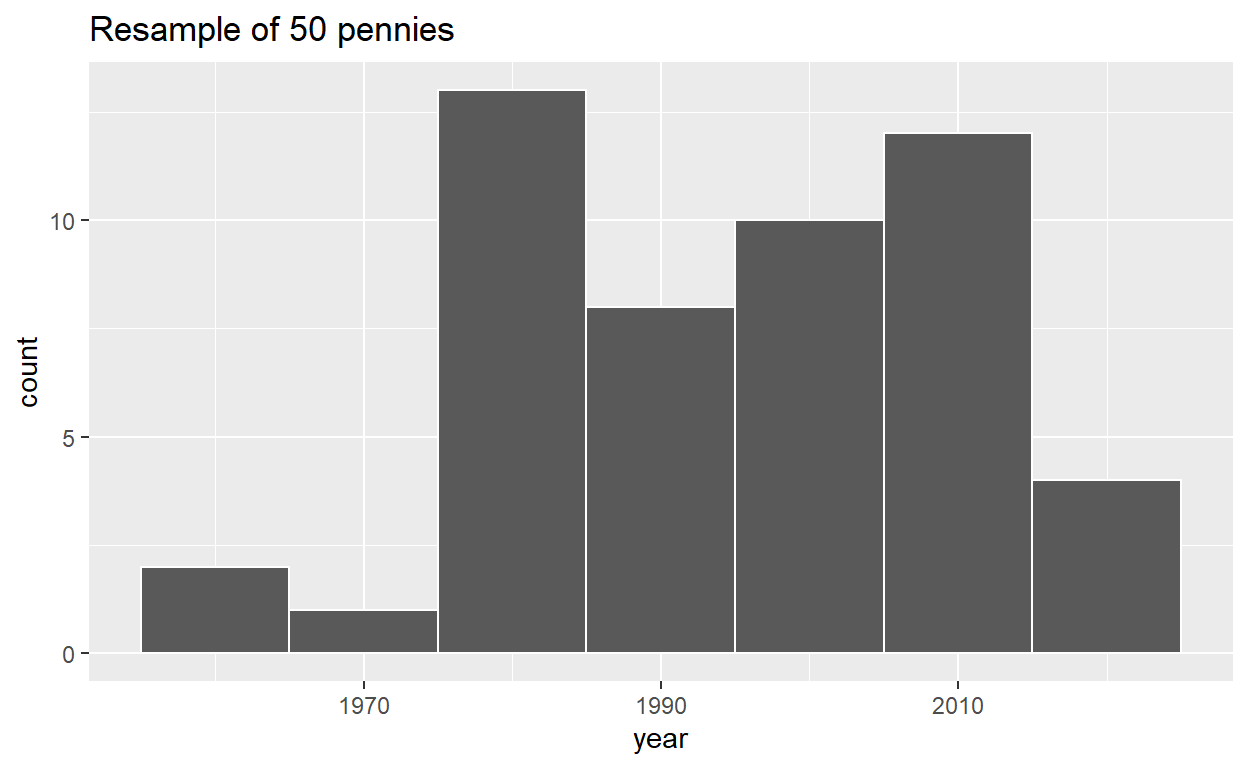
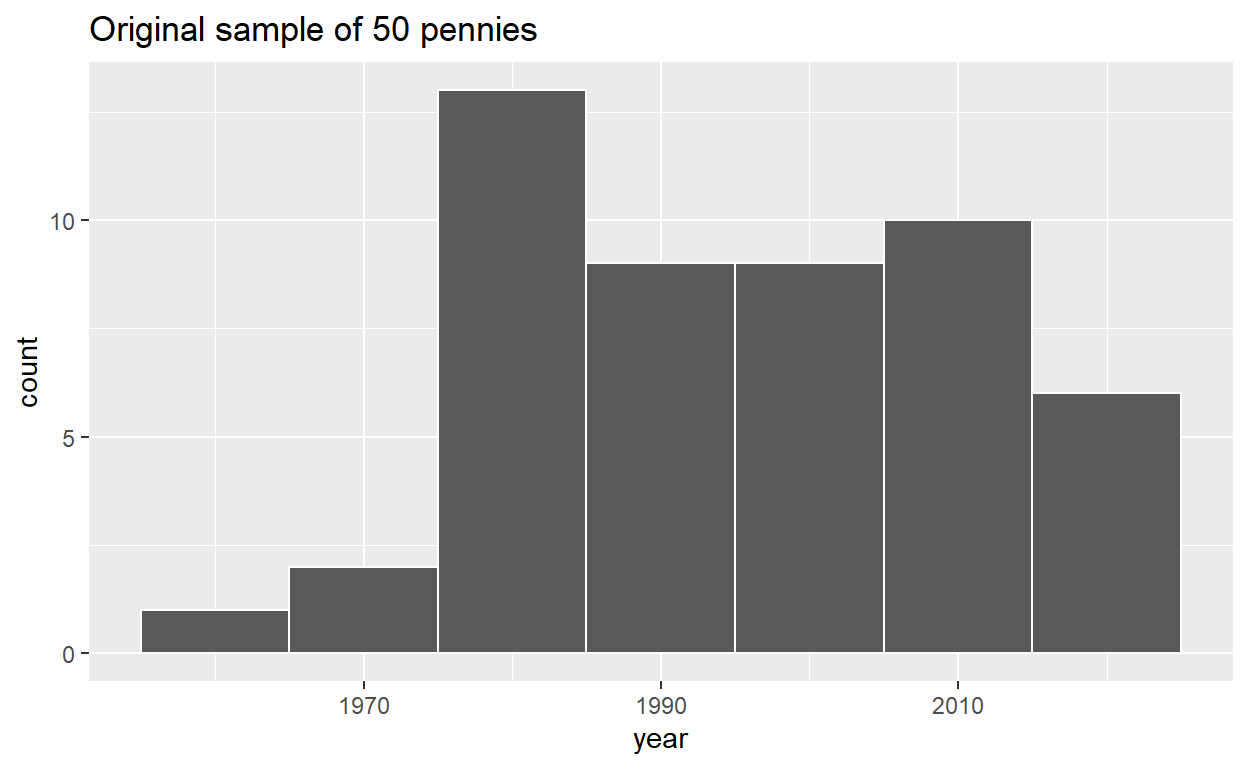
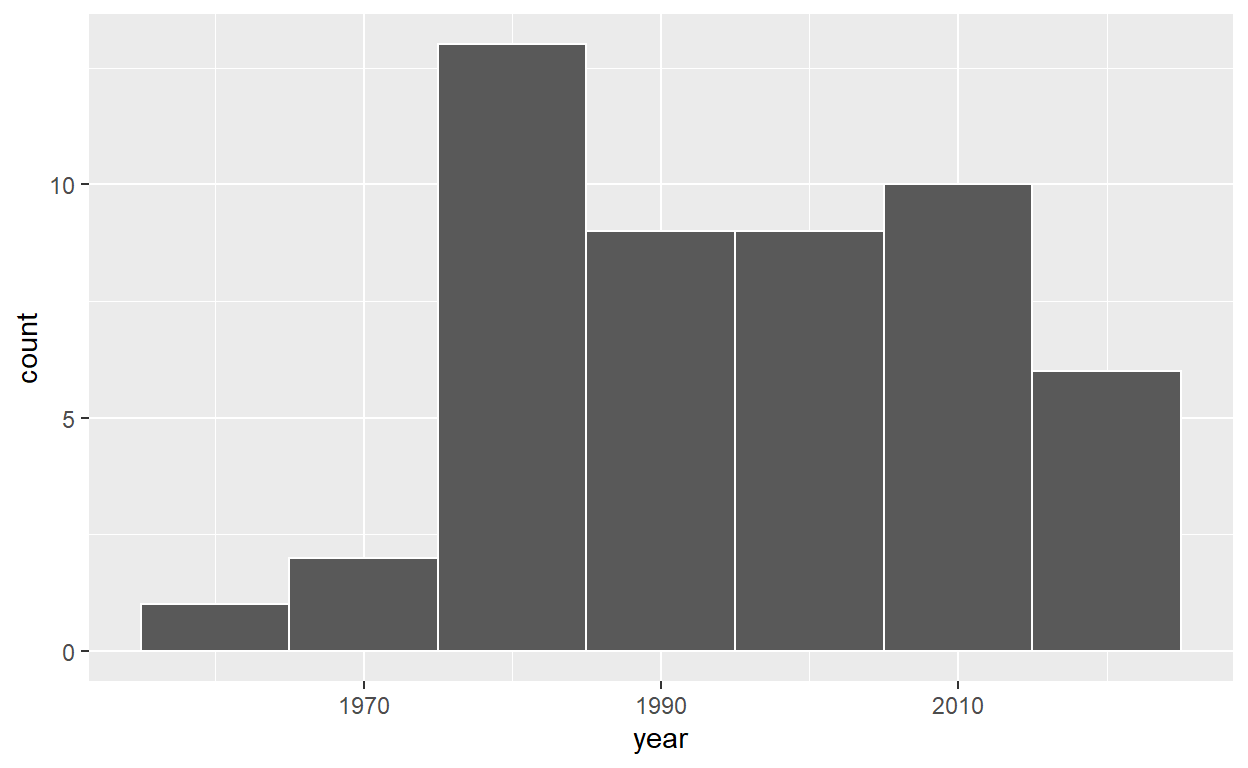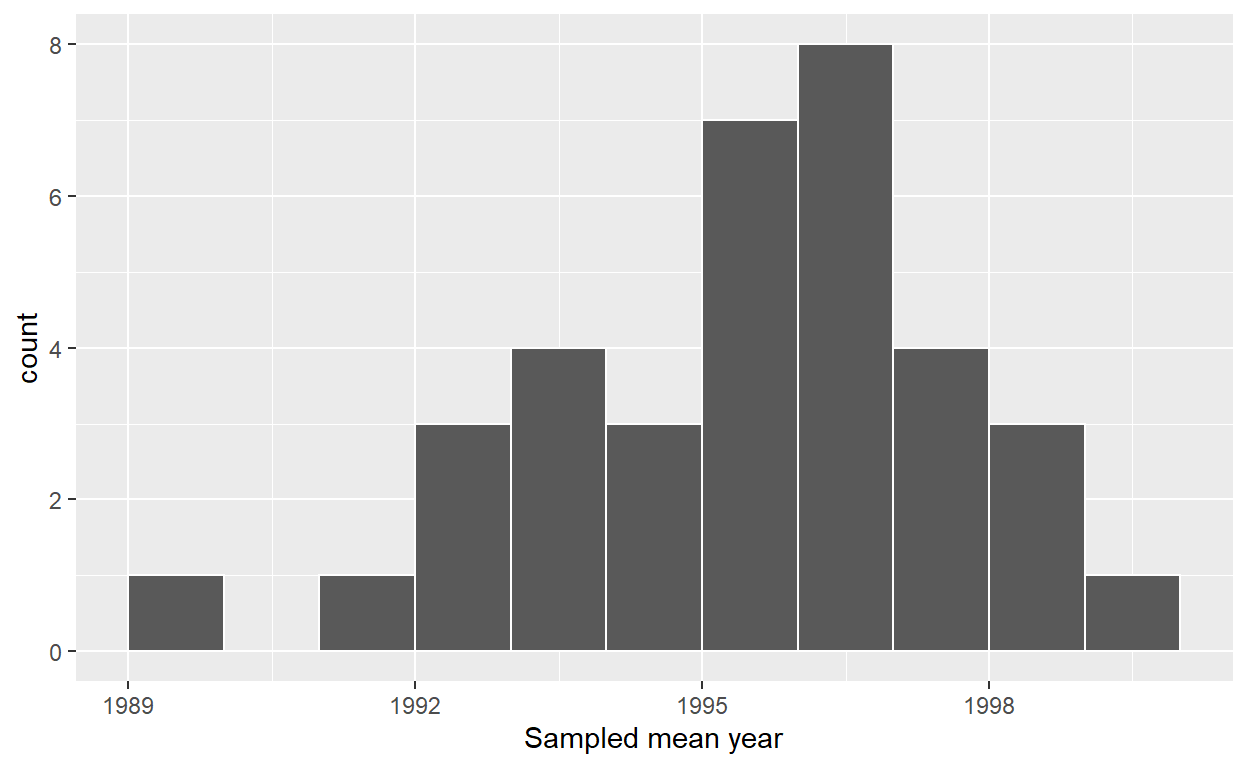# A tibble: 50 x 2
ID year
<int> <dbl>
1 1 2002
2 2 1986
3 3 2017
4 4 1988
5 5 2008
6 6 1983
7 7 2008
8 8 1996
9 9 2004
10 10 2000
# ... with 40 more rows
pennies_resample <- tibble(
year = c(1976, 1962, 1976, 1983, 2017, 2015, 2015, 1962, 2016, 1976,
2006, 1997, 1988, 2015, 2015, 1988, 2016, 1978, 1979, 1997,
1974, 2013, 1978, 2015, 2008, 1982, 1986, 1979, 1981, 2004,
2000, 1995, 1999, 2006, 1979, 2015, 1979, 1998, 1981, 2015,
2000, 1999, 1988, 2017, 1992, 1997, 1990, 1988, 2006, 2000)
)
# A tibble: 1 x 1
mean_year
<dbl>
1 1995.
# A tibble: 1,750 x 3
# Groups: name [35]
replicate name year
<int> <chr> <dbl>
1 1 Arianna 1988
2 1 Arianna 2002
3 1 Arianna 2015
4 1 Arianna 1998
5 1 Arianna 1979
6 1 Arianna 1971
7 1 Arianna 1971
8 1 Arianna 2015
9 1 Arianna 1988
10 1 Arianna 1979
# ... with 1,740 more rows
ggplot(resampled_means, aes(x = mean_year)) +
geom_histogram(binwidth = 1, color = "white", boundary = 1990) +
labs(x = "Sampled mean year")
virtual_resample <- pennies_sample %>%
rep_sample_n(size = 50, replace = TRUE)
# A tibble: 1 x 2
replicate resample_mean
<int> <dbl>
1 1 1997.
virtual_resamples <- pennies_sample %>%
rep_sample_n(size = 50, replace = TRUE, reps = 35)
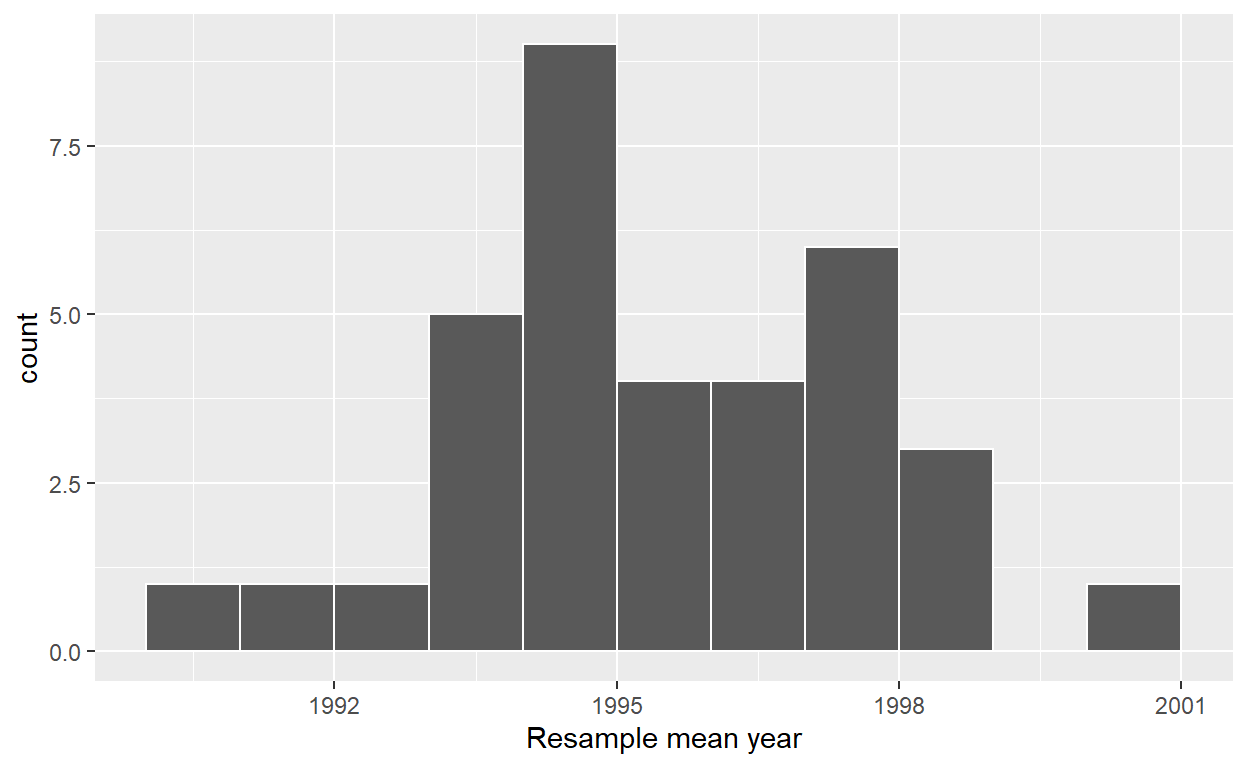
ggplot(virtual_resampled_means, aes(x = mean_year)) +
geom_histogram(binwidth = 1, color = "white", boundary = 1990) +
labs(x = "Resample mean year")
Repeat resampling 1000 times
virtual_resamples <- pennies_sample %>%
rep_sample_n(size = 50, replace = TRUE, reps = 1000)
Compute 1000 sample means
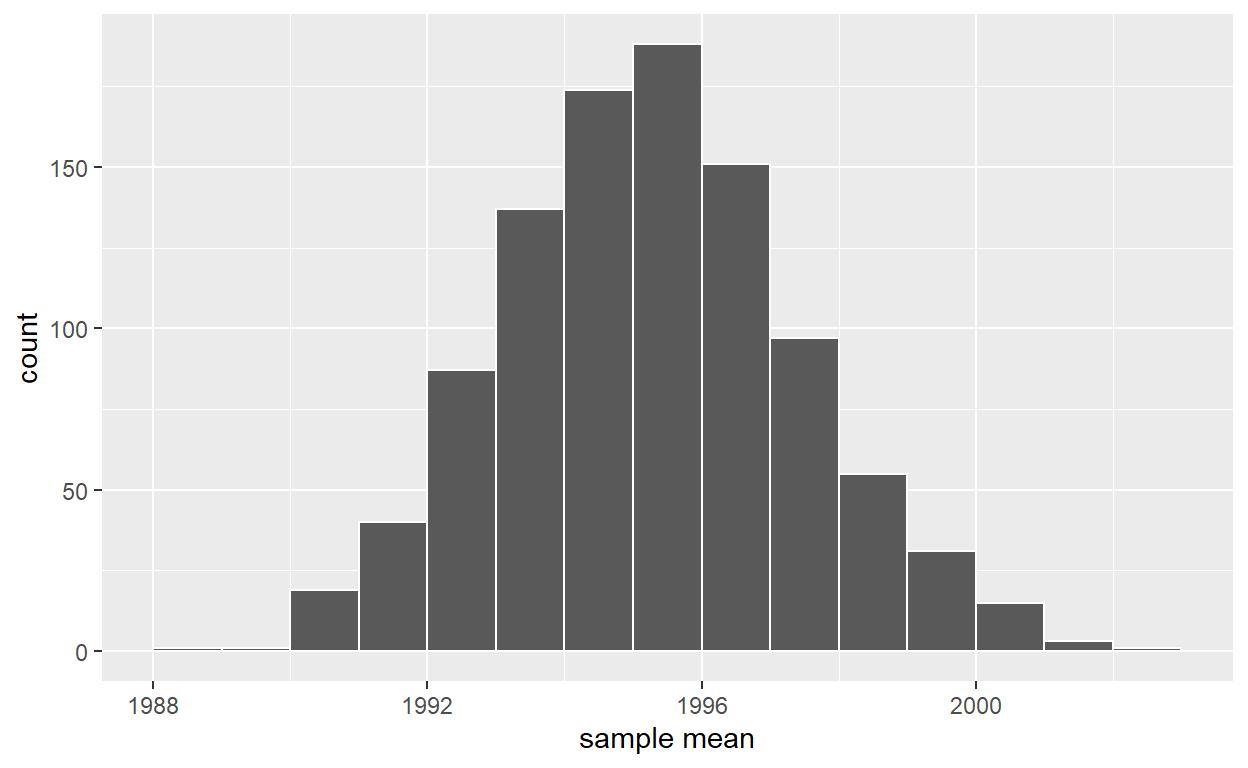
ggplot(virtual_resampled_means, aes(x = mean_year)) +
geom_histogram(binwidth = 1, color = "white", boundary = 1990) +
labs(x = "sample mean")
# A tibble: 1 x 1
mean_of_means
<dbl>
1 1995.
# A tibble: 1 x 1
SE
<dbl>
1 2.15
# A tibble: 50,000 x 3
# Groups: replicate [1,000]
replicate ID year
<int> <int> <dbl>
1 1 38 1999
2 1 47 1982
3 1 10 2000
4 1 12 1995
5 1 12 1995
6 1 50 2017
7 1 24 2017
8 1 3 2017
9 1 26 1979
10 1 15 1974
# ... with 49,990 more rows
# A tibble: 50,000 x 3
# Groups: replicate [1,000]
replicate ID year
<int> <int> <dbl>
1 1 12 1995
2 1 23 1998
3 1 6 1983
4 1 49 2006
5 1 33 1979
6 1 10 2000
7 1 27 1993
8 1 34 1985
9 1 35 1985
10 1 43 2018
# ... with 49,990 more rows
# A tibble: 1,000 x 2
replicate mean_year
<int> <dbl>
1 1 1996.
2 2 1989.
3 3 1996.
4 4 1995.
5 5 1998.
6 6 1997.
7 7 1994.
8 8 1995.
9 9 1995.
10 10 1996.
# ... with 990 more rows
# A tibble: 1 x 1
stat
<dbl>
1 1995.
Response: year (numeric)
# A tibble: 1 x 1
stat
<dbl>
1 1995.
Response: year (numeric)
# A tibble: 50 x 1
year
<dbl>
1 2002
2 1986
3 2017
4 1988
5 2008
6 1983
7 2008
8 1996
9 2004
10 2000
# ... with 40 more rows
Response: year (numeric)
# A tibble: 50 x 1
year
<dbl>
1 2002
2 1986
3 2017
4 1988
5 2008
6 1983
7 2008
8 1996
9 2004
10 2000
# ... with 40 more rows
Response: year (numeric)
# A tibble: 50,000 x 2
# Groups: replicate [1,000]
replicate year
<int> <dbl>
1 1 2004
2 1 1971
3 1 1996
4 1 1985
5 1 2006
6 1 1996
7 1 2015
8 1 1962
9 1 2017
10 1 1985
# ... with 49,990 more rows
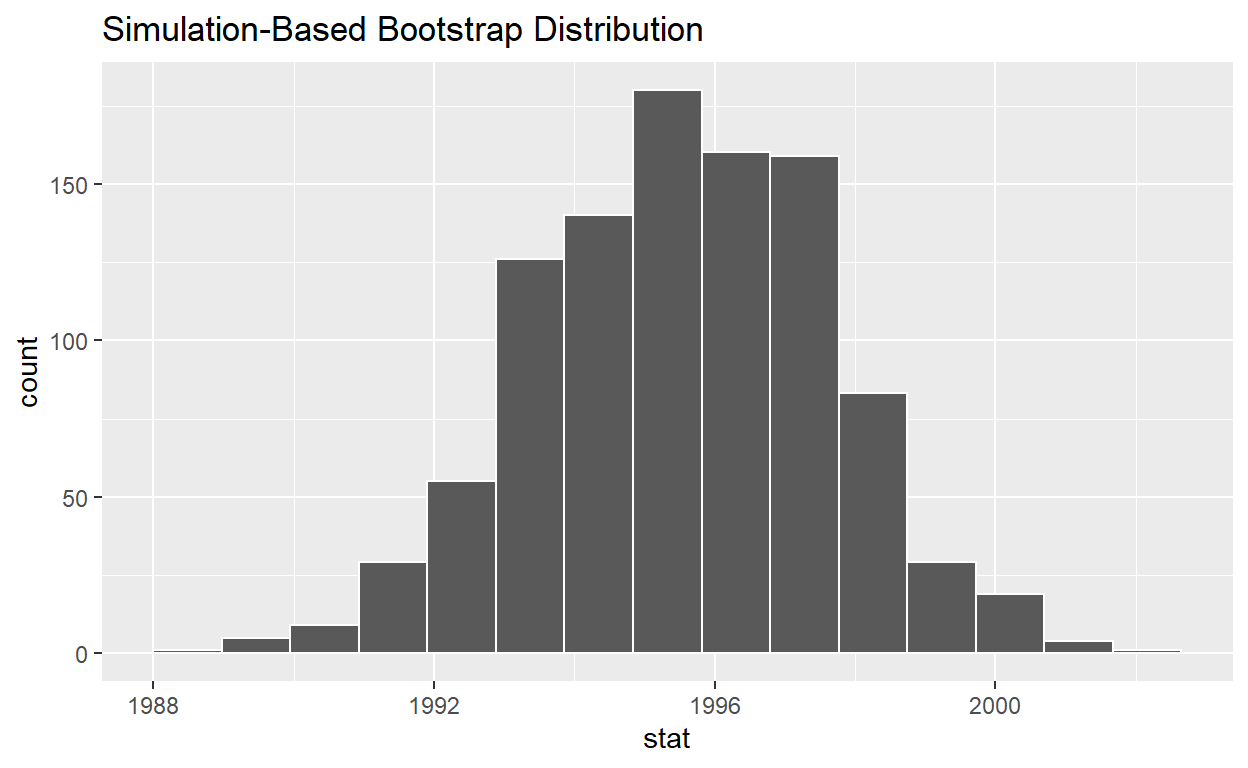
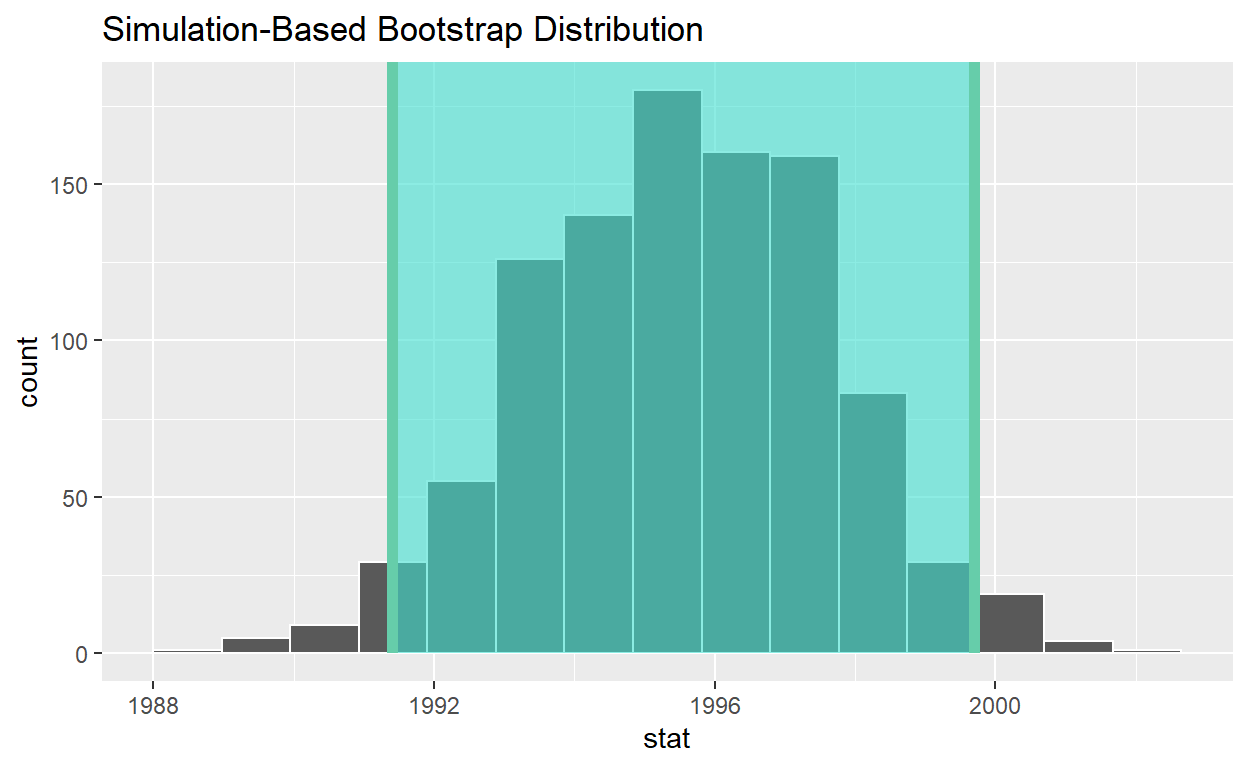
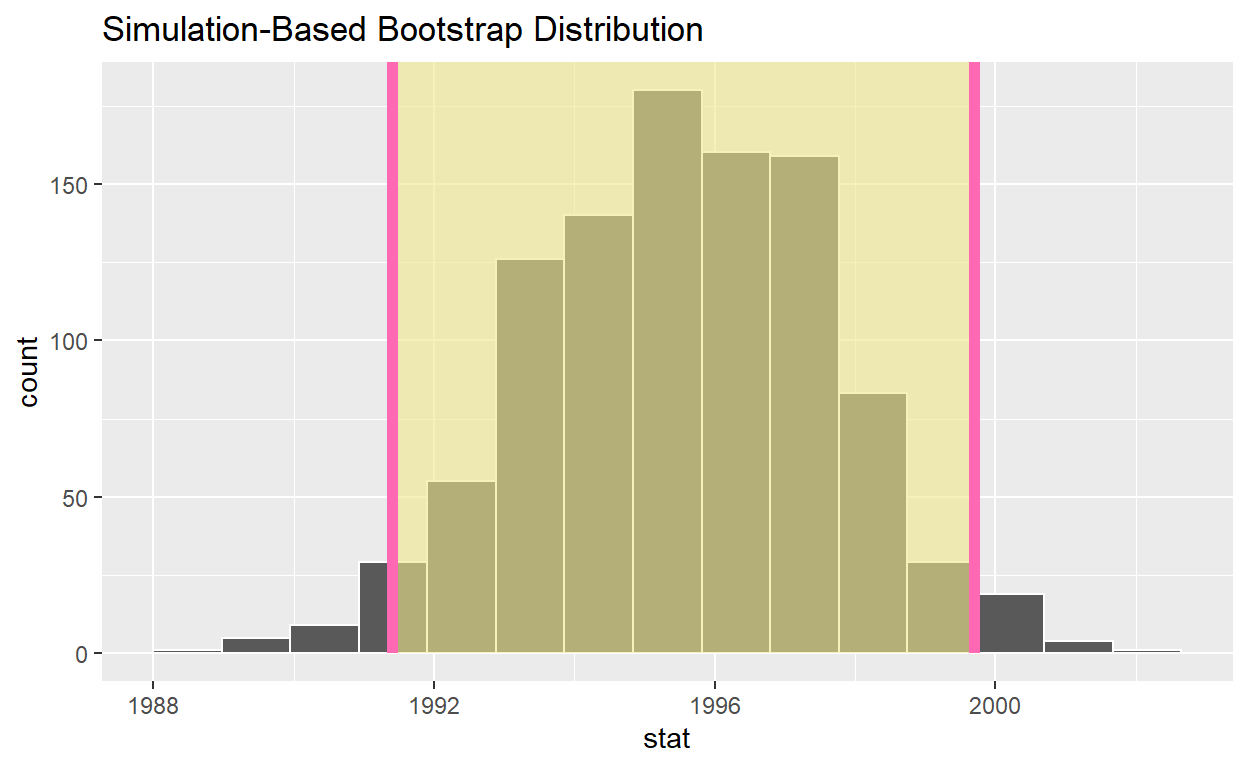
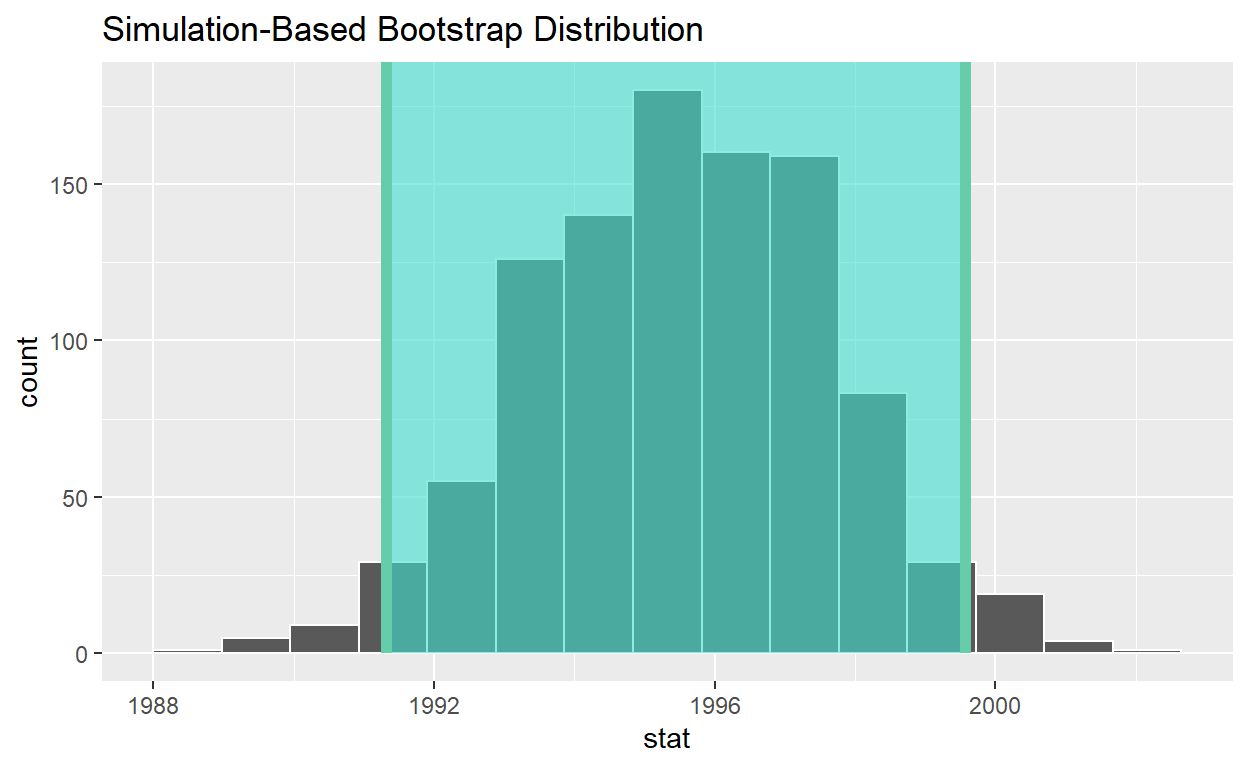
visualize(bootstrap_distribution) +
shade_ci(endpoints = percentile_ci, color = "hotpink", fill = "khaki")
# A tibble: 1 x 1
p_red
<dbl>
1 0.375
# A tibble: 50 x 1
color
<chr>
1 white
2 white
3 red
4 red
5 white
6 white
7 red
8 white
9 white
10 white
# ... with 40 more rows
bowl_sample_1 %>%
specify(response = color, success = "red")
Response: color (factor)
# A tibble: 50 x 1
color
<fct>
1 white
2 white
3 red
4 red
5 white
6 white
7 red
8 white
9 white
10 white
# ... with 40 more rows
bowl_sample_1 %>%
specify(response = color, success = "red") %>%
generate(reps = 1000, type = "bootstrap")
Response: color (factor)
# A tibble: 50,000 x 2
# Groups: replicate [1,000]
replicate color
<int> <fct>
1 1 white
2 1 white
3 1 red
4 1 red
5 1 white
6 1 white
7 1 white
8 1 red
9 1 white
10 1 red
# ... with 49,990 more rows
# A tibble: 50 x 3
subj group yawn
<int> <chr> <chr>
1 1 seed yes
2 2 control yes
3 3 seed no
4 4 seed yes
5 5 seed no
6 6 control no
7 7 seed yes
8 8 control no
9 9 control no
10 10 seed no
# ... with 40 more rows
# A tibble: 4 x 3
# Groups: group [2]
group yawn count
<chr> <chr> <int>
1 control no 12
2 control yes 4
3 seed no 24
4 seed yes 10
mythbusters_yawn %>%
specify(formula = yawn ~ group, success = "yes")
Response: yawn (factor)
Explanatory: group (factor)
# A tibble: 50 x 2
yawn group
<fct> <fct>
1 yes seed
2 yes control
3 no seed
4 yes seed
5 no seed
6 no control
7 yes seed
8 no control
9 no control
10 no seed
# ... with 40 more rows
first_six_rows <- head(mythbusters_yawn)
# A tibble: 6 x 3
subj group yawn
<int> <chr> <chr>
1 3 seed no
2 2 control yes
3 2 control yes
4 5 seed no
5 2 control yes
6 1 seed yes
mythbusters_yawn %>%
specify(formula = yawn ~ group, success = "yes") %>%
generate(reps = 1000, type = "bootstrap")
Response: yawn (factor)
Explanatory: group (factor)
# A tibble: 50,000 x 3
# Groups: replicate [1,000]
replicate yawn group
<int> <fct> <fct>
1 1 yes seed
2 1 no control
3 1 no seed
4 1 no seed
5 1 no seed
6 1 no seed
7 1 yes seed
8 1 yes seed
9 1 no seed
10 1 no control
# ... with 49,990 more rows
Response: yawn (factor)
Explanatory: group (factor)
# A tibble: 1,000 x 2
replicate stat
<int> <dbl>
1 1 -0.189
2 2 -0.373
3 3 0.0517
4 4 0.0333
5 5 -0.0921
6 6 0.0676
7 7 0.0167
8 8 -0.0963
9 9 -0.0164
10 10 0.190
# ... with 990 more rows
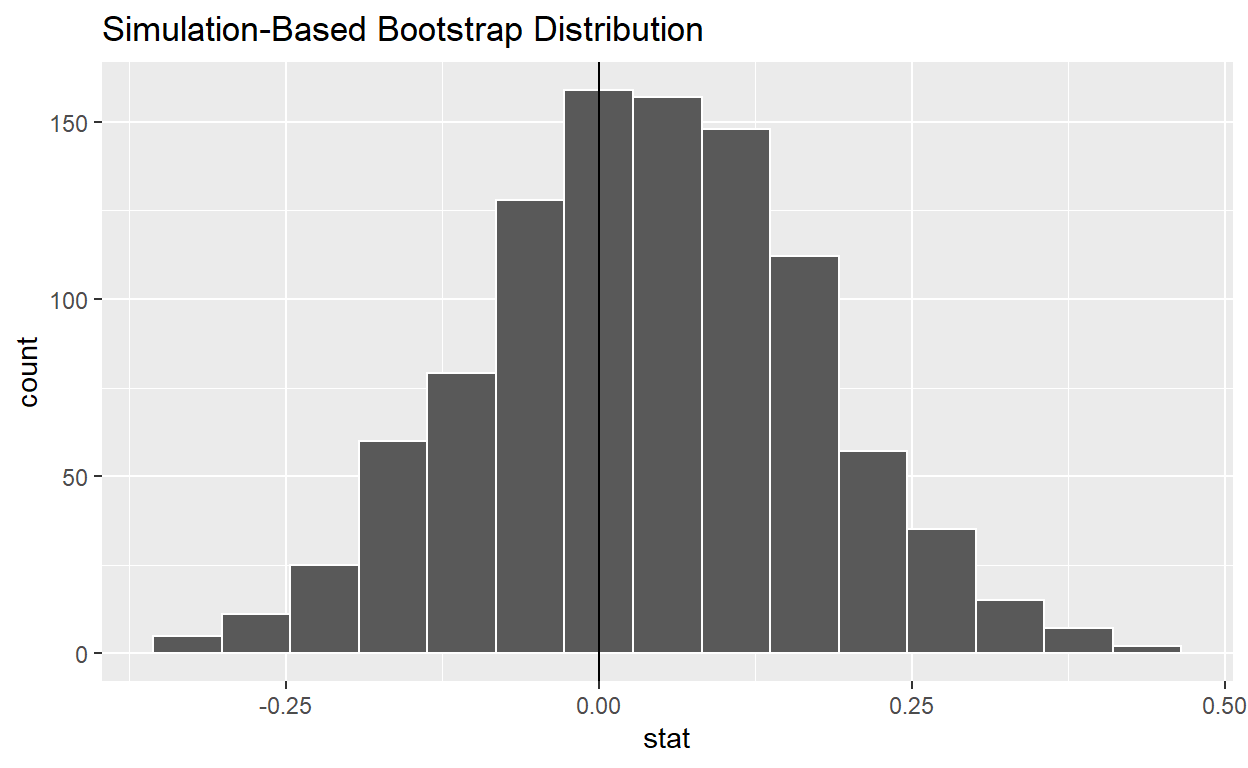
bootstrap_distribution_yawning <- mythbusters_yawn %>%
specify(formula = yawn ~ group, success = "yes") %>%
generate(reps = 1000, type = "bootstrap") %>%
calculate(stat = "diff in props", order = c("seed", "control"))
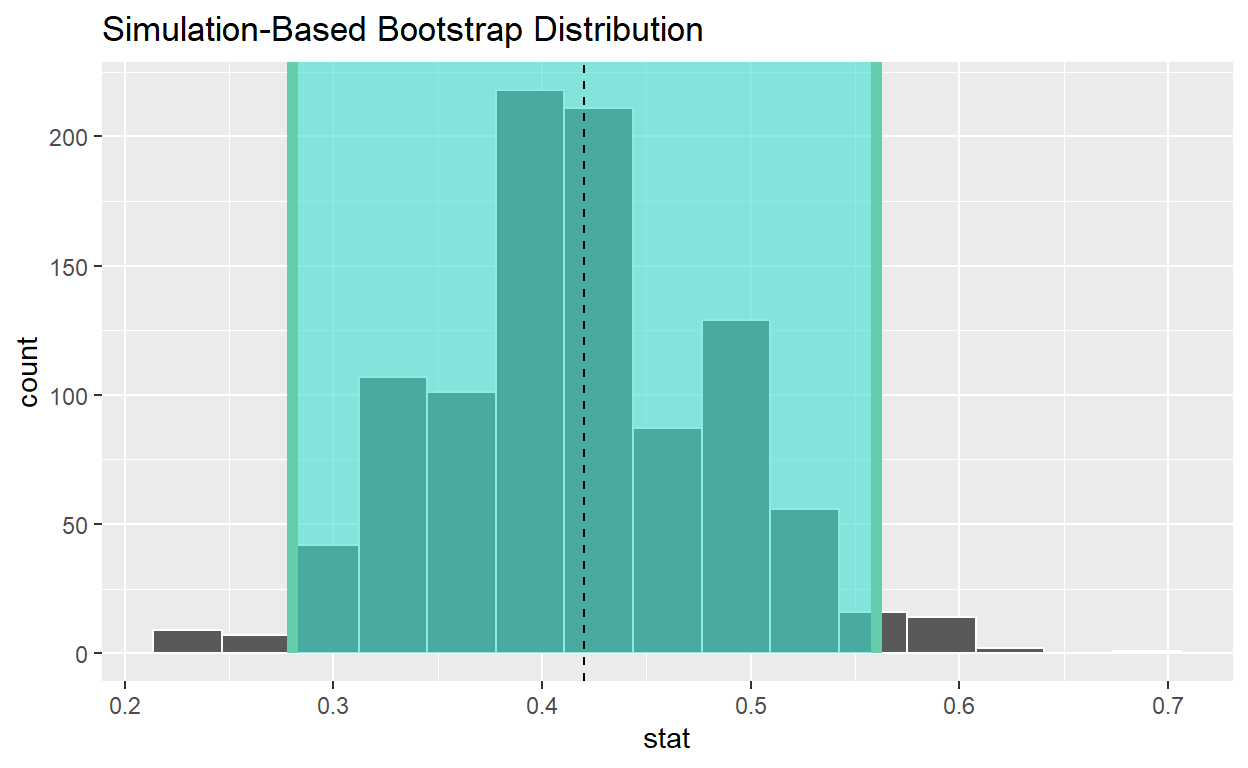
# A tibble: 1 x 2
lower_ci upper_ci
<dbl> <dbl>
1 -0.221 0.295
obs_diff_in_props <- mythbusters_yawn %>%
specify(formula = yawn ~ group, success = "yes") %>%
# generate(reps = 1000, type = "bootstrap") %>%
calculate(stat = "diff in props", order = c("seed", "control"))
# A tibble: 1 x 2
lower_ci upper_ci
<dbl> <dbl>
1 -0.215 0.304
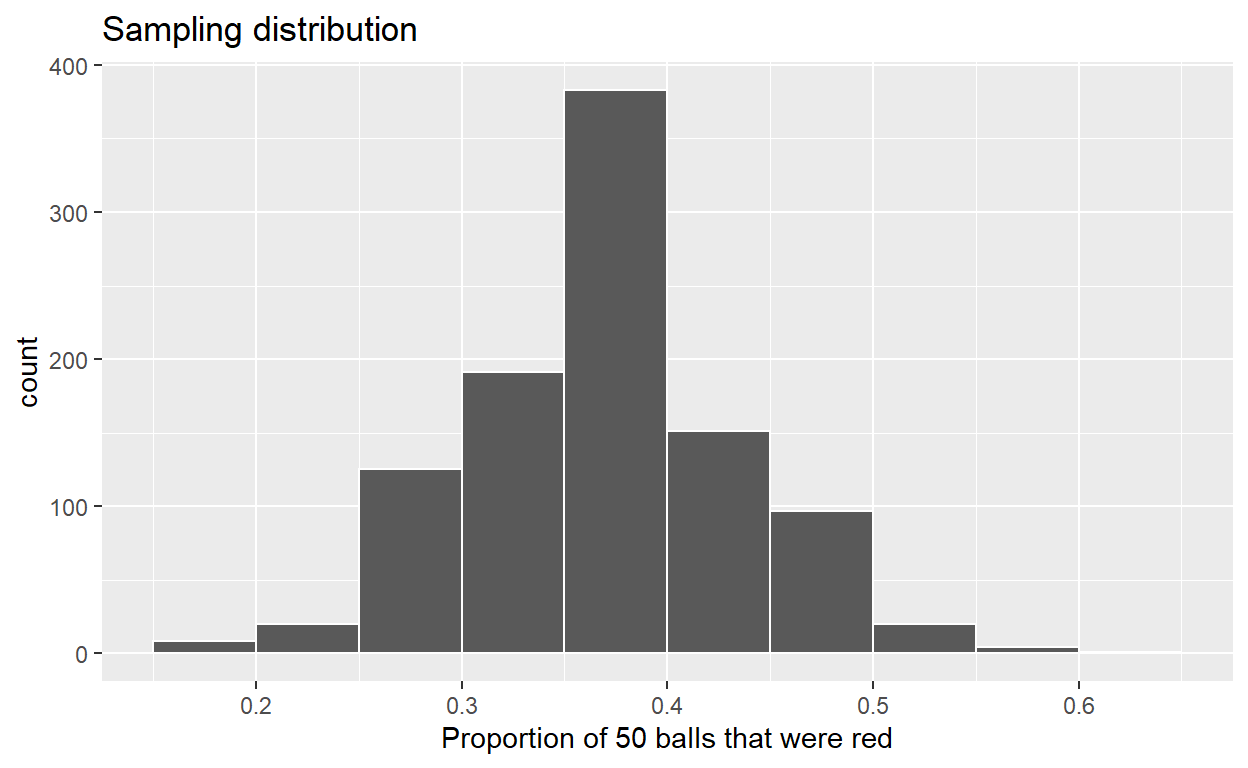
Take 1000 virtual samples of size 50 from the bowl:
Compute the sampling distribution of 1000 values of p-hat
Visualize sampling distribution of p-hat
ggplot(sampling_distribution, aes(x = prop_red)) +
geom_histogram(binwidth = 0.05, boundary = 0.4, color = "white") +
labs(x = "Proportion of 50 balls that were red",
title = "Sampling distribution")
# A tibble: 1 x 1
se
<dbl>
1 0.0658
# A tibble: 1 x 1
se
<dbl>
1 0.0711
conf_ints <- tactile_prop_red %>%
rename(p_hat = prop_red) %>%
mutate(
n = 50,
SE = sqrt(p_hat * (1 - p_hat) / n),
MoE = 1.96 * SE,
lower_ci = p_hat - MoE,
upper_ci = p_hat + MoE
)